1import pandas as pd- 1
-
Import the
pandaspackage under the aliaspd. Using the alias is not necessary, but it is a convention nearly everyone follows.
You know the basics. What are Jupyter notebooks, how do they work, and how do you run Python in them. It is time to start using them for data science (no, that simple math you did the last time doesn’t count as data science).
You are about to enter the PyData ecosystem. It means that you will start learning how to work with Python from the middle. This course does not explicitly cover the fundamentals of programming. It is expected that those parts you need you’ll be able to pick as you go through the specialised data science stack. If you’re stuck, confused or need further explanation, use Google (or your favourite search engine), ask AI to explain the code or ask in online chat or during the class. Not everything will be told during the course (by design), and the internet is a friend of every programmer, so let’s figure out how to use it efficiently from the beginning.
Let’s dig in!
Real-world datasets are messy. There is no way around it: datasets have “holes” (missing data), the amount of formats in which data can be stored is endless, and the best structure to share data is not always the optimum to analyse them, hence the need to munge1 them. As has been correctly pointed out in many outlets, much of the time spent in what is called Data Science is related not only to sophisticated modelling and insight but has to do with much more basic and less exotic tasks such as obtaining data, processing, and turning them into a shape that makes analysis possible, and exploring it to get to know their basic properties.
Surprisingly, very little has been published on patterns, techniques, and best practices for quick and efficient data cleaning, manipulation, and transformation because of how labour-intensive and relevant this aspect is. In this session, you will use a few real-world datasets and learn how to process them into Python so they can be transformed and manipulated, if necessary, and analysed. For this, you will introduce some of the bread and butter of data analysis and scientific computing in Python. These are fundamental tools that are constantly used in almost any task relating to data analysis.
This notebook covers the basics and the content that is expected to be learnt by every student. You use a prepared dataset that saves us much of the more intricate processing that goes beyond the introductory level the session is aimed at. If you are interested in how it was done, there is a notebook.
This notebook discusses several patterns to clean and structure data properly, including tidying, subsetting, and aggregating. You finish with some basic visualisation. An additional extension presents more advanced tricks to manipulate tabular data.
You will be exploring demographic characteristics of Chicago in 1918 linked to the influenza mortality during the pandemic that happened back then, coming from the research paper by Grantz et al. (2016). The data are aggregated to census tracts and contain information on unemployment, home ownership, age structure and influenza mortality from a period of 8 weeks.
The main tool you use is the pandas package. As with the math you used before, you must import it first.
1import pandas as pdpandas package under the alias pd. Using the alias is not necessary, but it is a convention nearly everyone follows.
The data is stored in a CSV file. To make things easier, you can read data from a file posted online so, for now, you do not need to download any dataset:
read_csv function from pandas. Remember that you have imported pandas as pd.
geography_code as an index of the table by passing its name to the index_col keyword argument. It is not strictly necessary but allows us to choose and index on reading instead of specifying it later. More on indices below.
You are using read_csv because the file you want to read is in CSV format. However, pandas allows for many more formats to be read and write. A full list of formats supported may be found in the documentation.
Instead of reading the file directly off the web, it is possible to download it manually, store it on your computer, and read it locally. To do that, you can follow these steps:
chicago_1918 = pd.read_csv(
"chicago_influenza_1918.csv",
index_col="geography_code",
)Now, you are ready to start playing and interrogating the dataset! What you have at your fingertips is a table summarising, for each of the census tracts in Chicago more than a century ago, how many people lived in each by age, accompanied by some other socioeconomic data and influenza mortality. These tables are called DataFrame objects, and they have a lot of functionality built-in to explore and manipulate the data they contain. Let’s explore a few of those cool tricks!
The first aspect worth spending a bit of time on is the structure of a DataFrame. You can print it by simply typing its name:
chicago_1918| gross_acres | illit | unemployed_pct | ho_pct | agecat1 | agecat2 | agecat3 | agecat4 | agecat5 | agecat6 | agecat7 | influenza | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| geography_code | ||||||||||||
| G17003100001 | 1388.2 | 116 | 0.376950 | 0.124823 | 46 | 274 | 257 | 311 | 222 | 1122 | 587 | 9 |
| G17003100002 | 217.7 | 14 | 0.399571 | 0.071647 | 35 | 320 | 441 | 624 | 276 | 1061 | 508 | 6 |
| G17003100003 | 401.3 | 69 | 0.349558 | 0.092920 | 50 | 265 | 179 | 187 | 163 | 1020 | 392 | 8 |
| G17003100004 | 86.9 | 11 | 0.422535 | 0.030072 | 43 | 241 | 129 | 141 | 123 | 1407 | 539 | 2 |
| G17003100005 | 337.1 | 20 | 0.431822 | 0.084703 | 65 | 464 | 369 | 464 | 328 | 2625 | 1213 | 7 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| G17003100492 | 2176.6 | 136 | 0.404430 | 0.173351 | 85 | 606 | 520 | 705 | 439 | 2141 | 1460 | 12 |
| G17003100493 | 680.0 | 271 | 0.377207 | 0.130158 | 243 | 1349 | 957 | 1264 | 957 | 4653 | 2180 | 40 |
| G17003100494 | 1392.8 | 1504 | 0.336032 | 0.072317 | 309 | 1779 | 1252 | 1598 | 1086 | 6235 | 2673 | 85 |
| G17003100495 | 640.0 | 167 | 0.311917 | 0.085667 | 59 | 333 | 206 | 193 | 80 | 726 | 224 | 15 |
| G17003100496 | 709.8 | 340 | 0.369765 | 0.113549 | 157 | 979 | 761 | 959 | 594 | 2862 | 1206 | 30 |
496 rows × 12 columns
Note the printing is cut to keep a nice and compact view but enough to see its structure. Since they represent a table of data, DataFrame objects have two dimensions: rows and columns. Each of these is automatically assigned a name in what you will call its index. When printing, the index of each dimension is rendered in bold, as opposed to the standard rendering for the content. The example above shows how the column index is automatically picked up from the .csv file’s column names. For rows, we have specified when reading the file you wanted the column geography_code, so that is used. If you hadn’t set any, pandas would automatically generate a sequence starting in 0 and going all the way to the number of rows minus one. This is the standard structure of a DataFrame object, so you will come to it over and over. Importantly, even when you move to spatial data, your datasets will have a similar structure.
One final feature that is worth mentioning about these tables is that they can hold columns with different types of data. In this example, you have counts (or int for integer types) and ratios (or ‘float’ for floating point numbers - a number with decimals) for each column. But it is useful to keep in mind that you can combine this with columns that hold other types of data such as categories, text (str, for string), dates or, as you will see later in the course, geographic features.
To extract a single column from this DataFrame, specify its name in the square brackets ([]). Note that the name, in this case, is a string. A piece of text. As such, it needs to be within single (') or double quotes ("). The resulting data structure is no longer a DataFrame, but you have a Series because you deal with a single column.
chicago_1918["influenza"]geography_code
G17003100001 9
G17003100002 6
G17003100003 8
G17003100004 2
G17003100005 7
..
G17003100492 12
G17003100493 40
G17003100494 85
G17003100495 15
G17003100496 30
Name: influenza, Length: 496, dtype: int64Inspecting what it looks like. You can check the table’s top (or bottom) X lines by passing X to the method head (tail). For example, for the top/bottom five lines:
chicago_1918.head()| gross_acres | illit | unemployed_pct | ho_pct | agecat1 | agecat2 | agecat3 | agecat4 | agecat5 | agecat6 | agecat7 | influenza | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| geography_code | ||||||||||||
| G17003100001 | 1388.2 | 116 | 0.376950 | 0.124823 | 46 | 274 | 257 | 311 | 222 | 1122 | 587 | 9 |
| G17003100002 | 217.7 | 14 | 0.399571 | 0.071647 | 35 | 320 | 441 | 624 | 276 | 1061 | 508 | 6 |
| G17003100003 | 401.3 | 69 | 0.349558 | 0.092920 | 50 | 265 | 179 | 187 | 163 | 1020 | 392 | 8 |
| G17003100004 | 86.9 | 11 | 0.422535 | 0.030072 | 43 | 241 | 129 | 141 | 123 | 1407 | 539 | 2 |
| G17003100005 | 337.1 | 20 | 0.431822 | 0.084703 | 65 | 464 | 369 | 464 | 328 | 2625 | 1213 | 7 |
chicago_1918.tail()| gross_acres | illit | unemployed_pct | ho_pct | agecat1 | agecat2 | agecat3 | agecat4 | agecat5 | agecat6 | agecat7 | influenza | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| geography_code | ||||||||||||
| G17003100492 | 2176.6 | 136 | 0.404430 | 0.173351 | 85 | 606 | 520 | 705 | 439 | 2141 | 1460 | 12 |
| G17003100493 | 680.0 | 271 | 0.377207 | 0.130158 | 243 | 1349 | 957 | 1264 | 957 | 4653 | 2180 | 40 |
| G17003100494 | 1392.8 | 1504 | 0.336032 | 0.072317 | 309 | 1779 | 1252 | 1598 | 1086 | 6235 | 2673 | 85 |
| G17003100495 | 640.0 | 167 | 0.311917 | 0.085667 | 59 | 333 | 206 | 193 | 80 | 726 | 224 | 15 |
| G17003100496 | 709.8 | 340 | 0.369765 | 0.113549 | 157 | 979 | 761 | 959 | 594 | 2862 | 1206 | 30 |
Or get an overview of the table:
chicago_1918.info()<class 'pandas.core.frame.DataFrame'>
Index: 496 entries, G17003100001 to G17003100496
Data columns (total 12 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 gross_acres 496 non-null float64
1 illit 496 non-null int64
2 unemployed_pct 496 non-null float64
3 ho_pct 496 non-null float64
4 agecat1 496 non-null int64
5 agecat2 496 non-null int64
6 agecat3 496 non-null int64
7 agecat4 496 non-null int64
8 agecat5 496 non-null int64
9 agecat6 496 non-null int64
10 agecat7 496 non-null int64
11 influenza 496 non-null int64
dtypes: float64(3), int64(9)
memory usage: 66.5+ KBOr of the values of the table:
chicago_1918.describe()| gross_acres | illit | unemployed_pct | ho_pct | agecat1 | agecat2 | agecat3 | agecat4 | agecat5 | agecat6 | agecat7 | influenza | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | 496.000000 | 496.000000 | 496.000000 | 496.000000 | 496.000000 | 496.000000 | 496.000000 | 496.000000 | 496.000000 | 496.000000 | 496.000000 | 496.000000 |
| mean | 233.245968 | 199.116935 | 0.345818 | 0.061174 | 102.370968 | 555.167339 | 406.560484 | 524.100806 | 416.044355 | 2361.582661 | 1052.681452 | 16.070565 |
| std | 391.630857 | 297.836201 | 0.050498 | 0.038189 | 78.677423 | 423.526444 | 301.564896 | 369.875444 | 281.825682 | 1545.469426 | 722.955717 | 12.252440 |
| min | 6.900000 | 0.000000 | 0.057800 | 0.000000 | 0.000000 | 3.000000 | 1.000000 | 4.000000 | 0.000000 | 8.000000 | 6.000000 | 0.000000 |
| 25% | 79.975000 | 30.750000 | 0.323973 | 0.032106 | 46.750000 | 256.500000 | 193.500000 | 253.750000 | 220.500000 | 1169.750000 | 519.750000 | 8.000000 |
| 50% | 99.500000 | 84.000000 | 0.353344 | 0.054389 | 82.000000 | 442.500000 | 331.500000 | 453.500000 | 377.000000 | 2102.000000 | 918.500000 | 13.500000 |
| 75% | 180.125000 | 241.250000 | 0.373382 | 0.084762 | 136.000000 | 717.500000 | 532.500000 | 709.500000 | 551.750000 | 3191.750000 | 1379.250000 | 21.000000 |
| max | 3840.000000 | 2596.000000 | 0.495413 | 0.197391 | 427.000000 | 2512.000000 | 1917.000000 | 2665.000000 | 2454.000000 | 9792.000000 | 4163.000000 | 85.000000 |
Note how the output is also a DataFrame object, so you can do with it the same things you would with the original table (e.g. writing it to a file).
In this case, the summary might be better presented if the table is “transposed”:
chicago_1918.describe().T| count | mean | std | min | 25% | 50% | 75% | max | |
|---|---|---|---|---|---|---|---|---|
| gross_acres | 496.0 | 233.245968 | 391.630857 | 6.9000 | 79.975000 | 99.500000 | 180.125000 | 3840.000000 |
| illit | 496.0 | 199.116935 | 297.836201 | 0.0000 | 30.750000 | 84.000000 | 241.250000 | 2596.000000 |
| unemployed_pct | 496.0 | 0.345818 | 0.050498 | 0.0578 | 0.323973 | 0.353344 | 0.373382 | 0.495413 |
| ho_pct | 496.0 | 0.061174 | 0.038189 | 0.0000 | 0.032106 | 0.054389 | 0.084762 | 0.197391 |
| agecat1 | 496.0 | 102.370968 | 78.677423 | 0.0000 | 46.750000 | 82.000000 | 136.000000 | 427.000000 |
| agecat2 | 496.0 | 555.167339 | 423.526444 | 3.0000 | 256.500000 | 442.500000 | 717.500000 | 2512.000000 |
| agecat3 | 496.0 | 406.560484 | 301.564896 | 1.0000 | 193.500000 | 331.500000 | 532.500000 | 1917.000000 |
| agecat4 | 496.0 | 524.100806 | 369.875444 | 4.0000 | 253.750000 | 453.500000 | 709.500000 | 2665.000000 |
| agecat5 | 496.0 | 416.044355 | 281.825682 | 0.0000 | 220.500000 | 377.000000 | 551.750000 | 2454.000000 |
| agecat6 | 496.0 | 2361.582661 | 1545.469426 | 8.0000 | 1169.750000 | 2102.000000 | 3191.750000 | 9792.000000 |
| agecat7 | 496.0 | 1052.681452 | 722.955717 | 6.0000 | 519.750000 | 918.500000 | 1379.250000 | 4163.000000 |
| influenza | 496.0 | 16.070565 | 12.252440 | 0.0000 | 8.000000 | 13.500000 | 21.000000 | 85.000000 |
Equally, common descriptive statistics are also available. To obtain minimum values for each column, you can use .min().
chicago_1918.min()gross_acres 6.9000
illit 0.0000
unemployed_pct 0.0578
ho_pct 0.0000
agecat1 0.0000
agecat2 3.0000
agecat3 1.0000
agecat4 4.0000
agecat5 0.0000
agecat6 8.0000
agecat7 6.0000
influenza 0.0000
dtype: float64Or to obtain a minimum for a single column only.
chicago_1918["influenza"].min()0Note here how you have restricted the calculation of the minimum value to one column only by getting the Series and calling .min() on that.
Similarly, you can restrict the calculations to a single row using .loc[] indexer:
chicago_1918.loc["G17003100492"].max()2176.6You can generate new variables by applying operations to existing ones. For example, you can calculate the total population by area. Here are a couple of ways to do it:
total_population to store the result.
geography_code
G17003100001 2819
G17003100002 3265
G17003100003 2256
G17003100004 2623
G17003100005 5528
dtype: int64# This one is shorted, using a range of columns and sum
1total_population = chicago_1918.loc[:, "agecat1":"agecat7"].sum(axis=1)
total_population.head().loc[], you select all the rows (: part) and all the columns between "agecat1" and "agecat7". Then you apply .sum() over axis=1, which means along rows, to get a sum per each row.
geography_code
G17003100001 2819
G17003100002 3265
G17003100003 2256
G17003100004 2623
G17003100005 5528
dtype: int64Once you have created the variable, you can make it part of the table:
1chicago_1918["total_population"] = total_population
chicago_1918.head()total_population that contains a Series as a column "total_population". pandas creates that column automatically. If it existed, it would get overridden.
| gross_acres | illit | unemployed_pct | ho_pct | agecat1 | agecat2 | agecat3 | agecat4 | agecat5 | agecat6 | agecat7 | influenza | total_population | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| geography_code | |||||||||||||
| G17003100001 | 1388.2 | 116 | 0.376950 | 0.124823 | 46 | 274 | 257 | 311 | 222 | 1122 | 587 | 9 | 2819 |
| G17003100002 | 217.7 | 14 | 0.399571 | 0.071647 | 35 | 320 | 441 | 624 | 276 | 1061 | 508 | 6 | 3265 |
| G17003100003 | 401.3 | 69 | 0.349558 | 0.092920 | 50 | 265 | 179 | 187 | 163 | 1020 | 392 | 8 | 2256 |
| G17003100004 | 86.9 | 11 | 0.422535 | 0.030072 | 43 | 241 | 129 | 141 | 123 | 1407 | 539 | 2 | 2623 |
| G17003100005 | 337.1 | 20 | 0.431822 | 0.084703 | 65 | 464 | 369 | 464 | 328 | 2625 | 1213 | 7 | 5528 |
You can also do other mathematical operations on columns. These are always automatically applied to individual values in corresponding rows.
1homeowners = chicago_1918["total_population"] * chicago_1918["ho_pct"]
homeowners.head()geography_code
G17003100001 351.875177
G17003100002 233.928353
G17003100003 209.628319
G17003100004 78.879711
G17003100005 468.237675
dtype: float641pop_density = chicago_1918["total_population"] / chicago_1918["gross_acres"]
pop_density.head()geography_code
G17003100001 2.030687
G17003100002 14.997703
G17003100003 5.621729
G17003100004 30.184120
G17003100005 16.398695
dtype: float64A different spin on this is assigning new values: you can generate new variables with scalars2, and modify those:
1chicago_1918["ones"] = 1
chicago_1918.head()"ones" with all ones.
| gross_acres | illit | unemployed_pct | ho_pct | agecat1 | agecat2 | agecat3 | agecat4 | agecat5 | agecat6 | agecat7 | influenza | total_population | ones | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| geography_code | ||||||||||||||
| G17003100001 | 1388.2 | 116 | 0.376950 | 0.124823 | 46 | 274 | 257 | 311 | 222 | 1122 | 587 | 9 | 2819 | 1 |
| G17003100002 | 217.7 | 14 | 0.399571 | 0.071647 | 35 | 320 | 441 | 624 | 276 | 1061 | 508 | 6 | 3265 | 1 |
| G17003100003 | 401.3 | 69 | 0.349558 | 0.092920 | 50 | 265 | 179 | 187 | 163 | 1020 | 392 | 8 | 2256 | 1 |
| G17003100004 | 86.9 | 11 | 0.422535 | 0.030072 | 43 | 241 | 129 | 141 | 123 | 1407 | 539 | 2 | 2623 | 1 |
| G17003100005 | 337.1 | 20 | 0.431822 | 0.084703 | 65 | 464 | 369 | 464 | 328 | 2625 | 1213 | 7 | 5528 | 1 |
And you can modify specific values too:
chicago_1918.loc["G17003100001", "ones"] = 3
chicago_1918.head()| gross_acres | illit | unemployed_pct | ho_pct | agecat1 | agecat2 | agecat3 | agecat4 | agecat5 | agecat6 | agecat7 | influenza | total_population | ones | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| geography_code | ||||||||||||||
| G17003100001 | 1388.2 | 116 | 0.376950 | 0.124823 | 46 | 274 | 257 | 311 | 222 | 1122 | 587 | 9 | 2819 | 3 |
| G17003100002 | 217.7 | 14 | 0.399571 | 0.071647 | 35 | 320 | 441 | 624 | 276 | 1061 | 508 | 6 | 3265 | 1 |
| G17003100003 | 401.3 | 69 | 0.349558 | 0.092920 | 50 | 265 | 179 | 187 | 163 | 1020 | 392 | 8 | 2256 | 1 |
| G17003100004 | 86.9 | 11 | 0.422535 | 0.030072 | 43 | 241 | 129 | 141 | 123 | 1407 | 539 | 2 | 2623 | 1 |
| G17003100005 | 337.1 | 20 | 0.431822 | 0.084703 | 65 | 464 | 369 | 464 | 328 | 2625 | 1213 | 7 | 5528 | 1 |
Permanently deleting variables is also within reach of one command:
chicago_1918 = chicago_1918.drop(columns="ones")
chicago_1918.head()| gross_acres | illit | unemployed_pct | ho_pct | agecat1 | agecat2 | agecat3 | agecat4 | agecat5 | agecat6 | agecat7 | influenza | total_population | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| geography_code | |||||||||||||
| G17003100001 | 1388.2 | 116 | 0.376950 | 0.124823 | 46 | 274 | 257 | 311 | 222 | 1122 | 587 | 9 | 2819 |
| G17003100002 | 217.7 | 14 | 0.399571 | 0.071647 | 35 | 320 | 441 | 624 | 276 | 1061 | 508 | 6 | 3265 |
| G17003100003 | 401.3 | 69 | 0.349558 | 0.092920 | 50 | 265 | 179 | 187 | 163 | 1020 | 392 | 8 | 2256 |
| G17003100004 | 86.9 | 11 | 0.422535 | 0.030072 | 43 | 241 | 129 | 141 | 123 | 1407 | 539 | 2 | 2623 |
| G17003100005 | 337.1 | 20 | 0.431822 | 0.084703 | 65 | 464 | 369 | 464 | 328 | 2625 | 1213 | 7 | 5528 |
Here, you explore how to subset parts of a DataFrame if you know exactly which bits you want. For example, if you want to extract the influenza mortality and total population of the first four areas in the table, you use loc with lists:
loc takes two inputs. Selection of rows and selection of columns. If the latter is not present, it is assumed that all the columns are selected. The same could be achieved by using :.
list of index values. Note that you use squared brackets ([]) to delineate the index of the items you want to subset. In Python, this sequence of items is called a list.
list of columns.
| influenza | total_population | |
|---|---|---|
| geography_code | ||
| G17003100001 | 9 | 2819 |
| G17003100002 | 6 | 3265 |
| G17003100003 | 8 | 2256 |
| G17003100004 | 2 | 2623 |
You can see how you can create a list with the names (index IDs) along each of the two dimensions of a DataFrame (rows and columns), and loc will return a subset of the original table only with the elements queried for.
An alternative to list-based queries is what is called “range-based” queries. These work on the indices of the table, but instead of requiring the ID of each item you want to retrieve, they operate by requiring only two IDs: the first and last element in a range of items. Range queries are expressed with a colon (:). For example:
range_query = chicago_1918.loc[
"G17003100010":"G17003100012",
"influenza":'total_population',
]
range_query| influenza | total_population | |
|---|---|---|
| geography_code | ||
| G17003100010 | 18 | 11380 |
| G17003100011 | 16 | 8382 |
| G17003100012 | 8 | 5874 |
The range query picks up all the elements between the specified IDs. Note that for this to work, the first ID in the range needs to be placed before the second one in the table’s index.
Once you know about list and range-based queries, you can combine them! For example, you can specify a range of rows and a list of columns:
range_list_qry = chicago_1918.loc[
"G17003100010":"G17003100012", ["influenza", "total_population"]
]
range_list_qry| influenza | total_population | |
|---|---|---|
| geography_code | ||
| G17003100010 | 18 | 11380 |
| G17003100011 | 16 | 8382 |
| G17003100012 | 8 | 5874 |
However, sometimes, you do not know exactly which observations you want, but you do know what conditions they need to satisfy (e.g. areas with more than 2,000 inhabitants). For these cases, DataFrames support selection based on conditions. Let us see a few examples. Suppose you want to select…
… areas with more than 60 cases of influenza deaths:
flu_over_60 = chicago_1918.loc[chicago_1918["influenza"] > 60]
flu_over_60| gross_acres | illit | unemployed_pct | ho_pct | agecat1 | agecat2 | agecat3 | agecat4 | agecat5 | agecat6 | agecat7 | influenza | total_population | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| geography_code | |||||||||||||
| G17003100196 | 80.5 | 951 | 0.301564 | 0.016648 | 228 | 1504 | 925 | 998 | 608 | 3583 | 1092 | 62 | 8938 |
| G17003100246 | 113.0 | 2596 | 0.330517 | 0.027537 | 407 | 2090 | 1497 | 1681 | 1063 | 4564 | 1934 | 77 | 13236 |
| G17003100247 | 91.1 | 1534 | 0.293748 | 0.020664 | 215 | 1136 | 770 | 775 | 530 | 2993 | 1152 | 61 | 7571 |
| G17003100285 | 120.0 | 621 | 0.318677 | 0.047779 | 419 | 1870 | 1190 | 1263 | 818 | 4370 | 1335 | 82 | 11265 |
| G17003100352 | 600.0 | 638 | 0.267962 | 0.055023 | 404 | 1962 | 1251 | 1151 | 603 | 5261 | 1251 | 70 | 11883 |
| G17003100494 | 1392.8 | 1504 | 0.336032 | 0.072317 | 309 | 1779 | 1252 | 1598 | 1086 | 6235 | 2673 | 85 | 14932 |
… areas with less than 200 inhabitants:
pop_under = chicago_1918.loc[chicago_1918["total_population"] < 200]
pop_under| gross_acres | illit | unemployed_pct | ho_pct | agecat1 | agecat2 | agecat3 | agecat4 | agecat5 | agecat6 | agecat7 | influenza | total_population | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| geography_code | |||||||||||||
| G17003100093 | 71.2 | 34 | 0.268041 | 0.025773 | 4 | 33 | 20 | 25 | 10 | 66 | 36 | 0 | 194 |
| G17003100293 | 40.0 | 17 | 0.345238 | 0.053571 | 4 | 23 | 18 | 26 | 16 | 59 | 22 | 0 | 168 |
| G17003100297 | 38.1 | 4 | 0.461538 | 0.000000 | 0 | 3 | 1 | 4 | 0 | 8 | 10 | 0 | 26 |
| G17003100353 | 320.0 | 28 | 0.193750 | 0.018750 | 1 | 7 | 4 | 10 | 12 | 80 | 46 | 2 | 160 |
| G17003100488 | 1600.1 | 3 | 0.404762 | 0.000000 | 2 | 5 | 4 | 7 | 1 | 17 | 6 | 0 | 42 |
… areas with exactly a hundred illiterate persons:
illit_100 = chicago_1918.loc[chicago_1918["illit"] == 100]
illit_100| gross_acres | illit | unemployed_pct | ho_pct | agecat1 | agecat2 | agecat3 | agecat4 | agecat5 | agecat6 | agecat7 | influenza | total_population | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| geography_code | |||||||||||||
| G17003100076 | 79.4 | 100 | 0.326936 | 0.043691 | 140 | 791 | 562 | 666 | 520 | 3163 | 1496 | 20 | 7338 |
| G17003100483 | 670.0 | 100 | 0.379153 | 0.142368 | 188 | 1170 | 829 | 1132 | 763 | 3191 | 1482 | 21 | 8755 |
These queries can grow in sophistication with almost no limits. For example, here is a case where you want to find out the areas where the oldest age group is more than half the population:
chicago_1918.loc[
(chicago_1918["agecat7"] * 100 / chicago_1918["total_population"]) > 50
]| gross_acres | illit | unemployed_pct | ho_pct | agecat1 | agecat2 | agecat3 | agecat4 | agecat5 | agecat6 | agecat7 | influenza | total_population | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| geography_code | |||||||||||||
| G17003100227 | 146.3 | 22 | 0.0657 | 0.000853 | 2 | 16 | 9 | 22 | 27 | 480 | 614 | 3 | 1170 |
All the condition-based queries above are expressed using the loc operator. This is a powerful way, and since it shares syntax with index-based queries, it is also easier to remember. However, sometimes querying using loc involves a lot of quotation marks, parenthesis, etc. A more streamlined approach for condition-based queries of rows is provided by the query engine. Using this approach, you express everything in our query on a single string, or piece of text, and that is evaluated in the table at once. For example, you can run the same operation as in the first query above with the following syntax:
flu_over_60_query = chicago_1918.query("influenza > 60")
flu_over_60_query| gross_acres | illit | unemployed_pct | ho_pct | agecat1 | agecat2 | agecat3 | agecat4 | agecat5 | agecat6 | agecat7 | influenza | total_population | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| geography_code | |||||||||||||
| G17003100196 | 80.5 | 951 | 0.301564 | 0.016648 | 228 | 1504 | 925 | 998 | 608 | 3583 | 1092 | 62 | 8938 |
| G17003100246 | 113.0 | 2596 | 0.330517 | 0.027537 | 407 | 2090 | 1497 | 1681 | 1063 | 4564 | 1934 | 77 | 13236 |
| G17003100247 | 91.1 | 1534 | 0.293748 | 0.020664 | 215 | 1136 | 770 | 775 | 530 | 2993 | 1152 | 61 | 7571 |
| G17003100285 | 120.0 | 621 | 0.318677 | 0.047779 | 419 | 1870 | 1190 | 1263 | 818 | 4370 | 1335 | 82 | 11265 |
| G17003100352 | 600.0 | 638 | 0.267962 | 0.055023 | 404 | 1962 | 1251 | 1151 | 603 | 5261 | 1251 | 70 | 11883 |
| G17003100494 | 1392.8 | 1504 | 0.336032 | 0.072317 | 309 | 1779 | 1252 | 1598 | 1086 | 6235 | 2673 | 85 | 14932 |
If you want to combine operations, this is also possible:
flu_query = chicago_1918.query("(influenza > 60) & (total_population < 10000)")
flu_query| gross_acres | illit | unemployed_pct | ho_pct | agecat1 | agecat2 | agecat3 | agecat4 | agecat5 | agecat6 | agecat7 | influenza | total_population | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| geography_code | |||||||||||||
| G17003100196 | 80.5 | 951 | 0.301564 | 0.016648 | 228 | 1504 | 925 | 998 | 608 | 3583 | 1092 | 62 | 8938 |
| G17003100247 | 91.1 | 1534 | 0.293748 | 0.020664 | 215 | 1136 | 770 | 775 | 530 | 2993 | 1152 | 61 | 7571 |
Note that, in these cases, using query results in code that is much more streamlined and easier to read. However, query is not perfect and, particularly for more sophisticated queries, it does not afford the same degree of flexibility. For example, the last query we had using loc would not be possible using query.
If you are interested, more detail about query is available in the pandas documentation.
Now, all of these queries can be combined with each other for further flexibility. For example, imagine you want areas with more than 60 cases of influenza from areas with less than 10,000 inhabitants:
flu_loc = chicago_1918.loc[
(chicago_1918["influenza"] > 60)
1 & (chicago_1918["total_population"] < 10000)
]
flu_loc& operator combines both conditions together.
| gross_acres | illit | unemployed_pct | ho_pct | agecat1 | agecat2 | agecat3 | agecat4 | agecat5 | agecat6 | agecat7 | influenza | total_population | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| geography_code | |||||||||||||
| G17003100196 | 80.5 | 951 | 0.301564 | 0.016648 | 228 | 1504 | 925 | 998 | 608 | 3583 | 1092 | 62 | 8938 |
| G17003100247 | 91.1 | 1534 | 0.293748 | 0.020664 | 215 | 1136 | 770 | 775 | 530 | 2993 | 1152 | 61 | 7571 |
loc queries work?
Let’s unpack how these queries work. Each part of the query above creates a single Series with boolean (True or False) values, encoding whether the row fulfils the condition or not.
chicago_1918["influenza"] > 60geography_code
G17003100001 False
G17003100002 False
G17003100003 False
G17003100004 False
G17003100005 False
...
G17003100492 False
G17003100493 False
G17003100494 True
G17003100495 False
G17003100496 False
Name: influenza, Length: 496, dtype: boolchicago_1918["total_population"] < 10000geography_code
G17003100001 True
G17003100002 True
G17003100003 True
G17003100004 True
G17003100005 True
...
G17003100492 True
G17003100493 False
G17003100494 False
G17003100495 True
G17003100496 True
Name: total_population, Length: 496, dtype: boolYou then combine two of these Series with &, asking for a new Series where values in both the first and the second Series are True.
(chicago_1918["influenza"] > 60) & (chicago_1918["total_population"] < 10000)geography_code
G17003100001 False
G17003100002 False
G17003100003 False
G17003100004 False
G17003100005 False
...
G17003100492 False
G17003100493 False
G17003100494 False
G17003100495 False
G17003100496 False
Length: 496, dtype: boolSuch a Series is then essentially used as a mask, and loc returns only those columns that contain True in that mask.
Among the many operations DataFrame objects support, one of the most useful ones is to sort a table based on a given column. For example, imagine you want to sort the table by the influenza cases:
1chicago_sorted = chicago_1918.sort_values('influenza', ascending=False)
chicago_sortedpandas is sorting from the smallest to the largest values (ascending). By specifying ascending=False, you switch the order.
| gross_acres | illit | unemployed_pct | ho_pct | agecat1 | agecat2 | agecat3 | agecat4 | agecat5 | agecat6 | agecat7 | influenza | total_population | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| geography_code | |||||||||||||
| G17003100494 | 1392.8 | 1504 | 0.336032 | 0.072317 | 309 | 1779 | 1252 | 1598 | 1086 | 6235 | 2673 | 85 | 14932 |
| G17003100285 | 120.0 | 621 | 0.318677 | 0.047779 | 419 | 1870 | 1190 | 1263 | 818 | 4370 | 1335 | 82 | 11265 |
| G17003100246 | 113.0 | 2596 | 0.330517 | 0.027537 | 407 | 2090 | 1497 | 1681 | 1063 | 4564 | 1934 | 77 | 13236 |
| G17003100352 | 600.0 | 638 | 0.267962 | 0.055023 | 404 | 1962 | 1251 | 1151 | 603 | 5261 | 1251 | 70 | 11883 |
| G17003100196 | 80.5 | 951 | 0.301564 | 0.016648 | 228 | 1504 | 925 | 998 | 608 | 3583 | 1092 | 62 | 8938 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| G17003100396 | 26.2 | 8 | 0.344066 | 0.015639 | 14 | 54 | 31 | 53 | 48 | 609 | 275 | 0 | 1084 |
| G17003100293 | 40.0 | 17 | 0.345238 | 0.053571 | 4 | 23 | 18 | 26 | 16 | 59 | 22 | 0 | 168 |
| G17003100488 | 1600.1 | 3 | 0.404762 | 0.000000 | 2 | 5 | 4 | 7 | 1 | 17 | 6 | 0 | 42 |
| G17003100297 | 38.1 | 4 | 0.461538 | 0.000000 | 0 | 3 | 1 | 4 | 0 | 8 | 10 | 0 | 26 |
| G17003100093 | 71.2 | 34 | 0.268041 | 0.025773 | 4 | 33 | 20 | 25 | 10 | 66 | 36 | 0 | 194 |
496 rows × 13 columns
Given the areas of each census tract differ, it may be better to sort by the mortality rate rather than raw counts.
| gross_acres | illit | unemployed_pct | ho_pct | agecat1 | agecat2 | agecat3 | agecat4 | agecat5 | agecat6 | agecat7 | influenza | total_population | flu_rate | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| geography_code | ||||||||||||||
| G17003100086 | 140.7 | 153 | 0.331750 | 0.034838 | 39 | 179 | 144 | 197 | 111 | 399 | 189 | 26 | 1258 | 0.020668 |
| G17003100022 | 233.2 | 20 | 0.369973 | 0.107239 | 6 | 28 | 35 | 48 | 28 | 151 | 77 | 7 | 373 | 0.018767 |
| G17003100261 | 164.9 | 287 | 0.307767 | 0.031068 | 25 | 126 | 92 | 113 | 87 | 414 | 172 | 18 | 1029 | 0.017493 |
| G17003100282 | 293.5 | 97 | 0.142330 | 0.044248 | 15 | 70 | 67 | 74 | 198 | 758 | 173 | 20 | 1355 | 0.014760 |
| G17003100249 | 137.0 | 317 | 0.337257 | 0.017202 | 63 | 314 | 277 | 345 | 171 | 718 | 320 | 31 | 2208 | 0.014040 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| G17003100396 | 26.2 | 8 | 0.344066 | 0.015639 | 14 | 54 | 31 | 53 | 48 | 609 | 275 | 0 | 1084 | 0.000000 |
| G17003100297 | 38.1 | 4 | 0.461538 | 0.000000 | 0 | 3 | 1 | 4 | 0 | 8 | 10 | 0 | 26 | 0.000000 |
| G17003100293 | 40.0 | 17 | 0.345238 | 0.053571 | 4 | 23 | 18 | 26 | 16 | 59 | 22 | 0 | 168 | 0.000000 |
| G17003100209 | 124.1 | 13 | 0.438105 | 0.062971 | 30 | 120 | 92 | 196 | 166 | 763 | 491 | 0 | 1858 | 0.000000 |
| G17003100202 | 13.1 | 6 | 0.323326 | 0.027714 | 5 | 40 | 24 | 37 | 28 | 218 | 78 | 0 | 430 | 0.000000 |
496 rows × 14 columns
If you inspect the help of chicago_1918.sort_values, you will find that you can pass more than one column to sort the table by. This allows you to do so-called hierarchical sorting: sort first based on one column, if equal, then based on another column, etc.
The next step to continue exploring a dataset is to get a feel for what it looks like, visually. You have already learnt how to unconver and inspect specific parts of the data, to check for particular cases you might be interested in. Now, you will see how to plot the data to get a sense of the overall distribution of values. For that, you can use the plotting capabilities of pandas.
One of the most common graphical devices to display the distribution of values in a variable is a histogram. Values are assigned into groups of equal intervals, and the groups are plotted as bars rising as high as the number of values into the group.
A histogram is easily created with the following command. In this case, let us have a look at the shape of the overall influenza rates:
_ = chicago_1918["influenza"].plot.hist()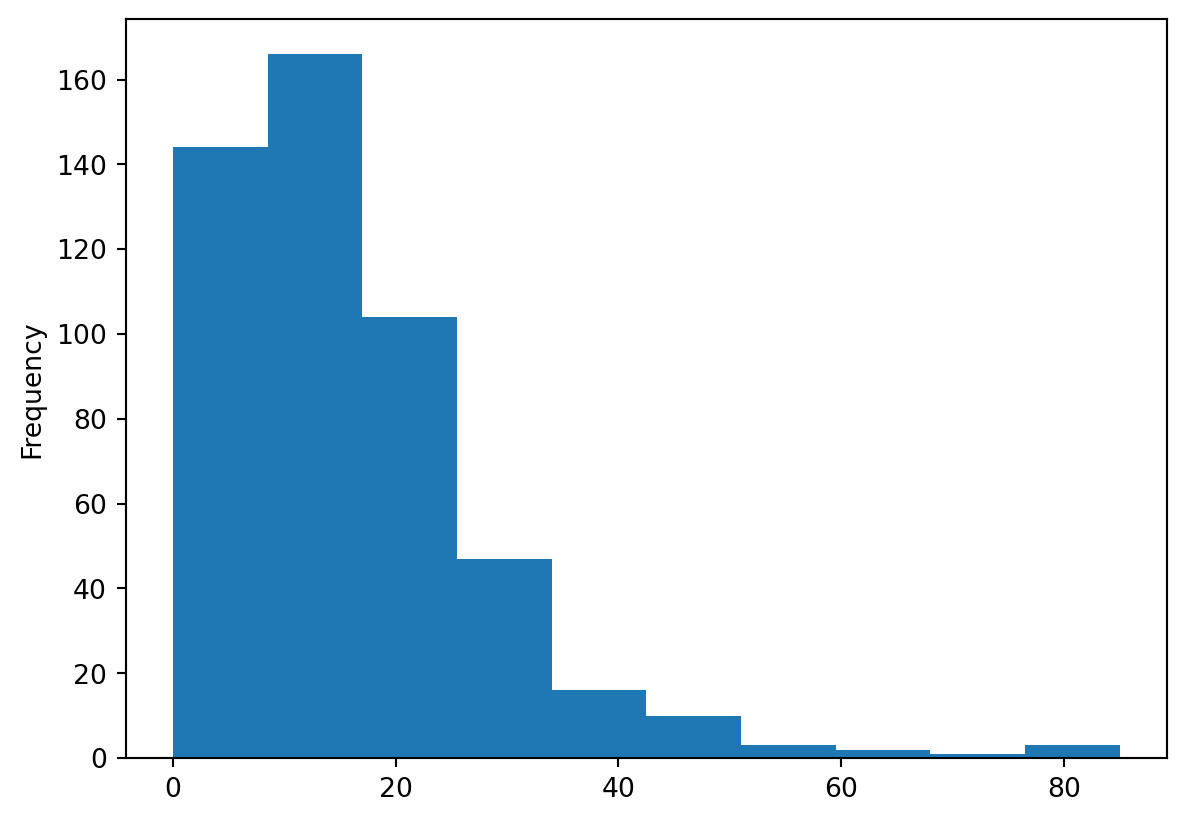
_
pandas returns an object with the drawing from its plotting methods. Since you are in Jupyter environment, and you don’t need to work further with that object; you can assign it to _, a convention for an unused variable.
However, the default pandas plots can be a bit dull. A better option is to use another package, called seaborn.
import seaborn as snssns?
seaborn is, by convention, imported as sns. That came as a joke after Samuel Normal Seaborn, a fictional character The West Wing show.
The same plot using seaborn has a more pleasant default style and more customisability.
sns.displot(chicago_1918["influenza"])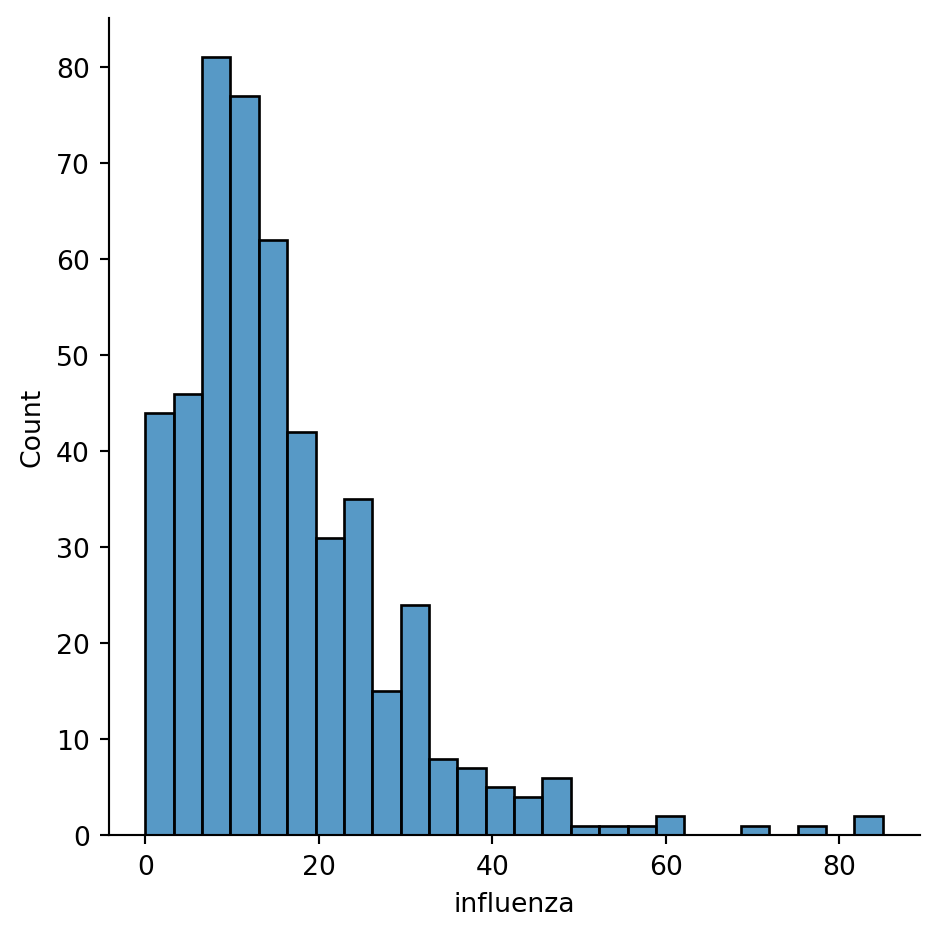
seabornNote you are using sns instead of pd, as the function belongs to seaborn instead of pandas.
You can quickly see most of the areas have seen somewhere between 0 and 60 cases, approx. However, there are a few areas that have more, up to more than 80 cases.
Histograms are useful, but they are artificial in the sense that a continuous variable is made discrete by turning the values into discrete groups. An alternative is kernel density estimation (KDE), which produces an empirical density function:
1sns.displot(chicago_1918["influenza"], kind="kde", fill=True)kind="kde" specifies which type of a distribution plot should seaborn use and fill=True tells it to colour the area under the KDE curve.
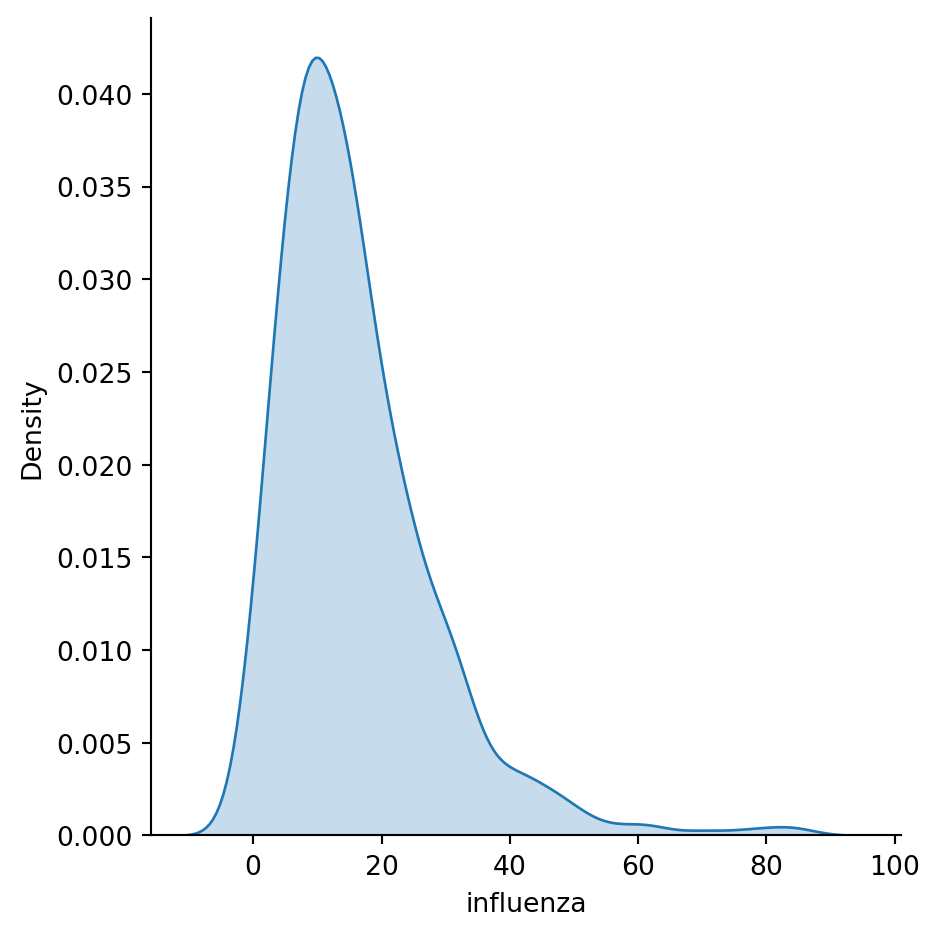
Another very common way of visually displaying a variable is with a line or a bar chart. For example, if you want to generate a line plot of the (sorted) total cases by area:
_ = chicago_1918["influenza"].sort_values(ascending=False).plot()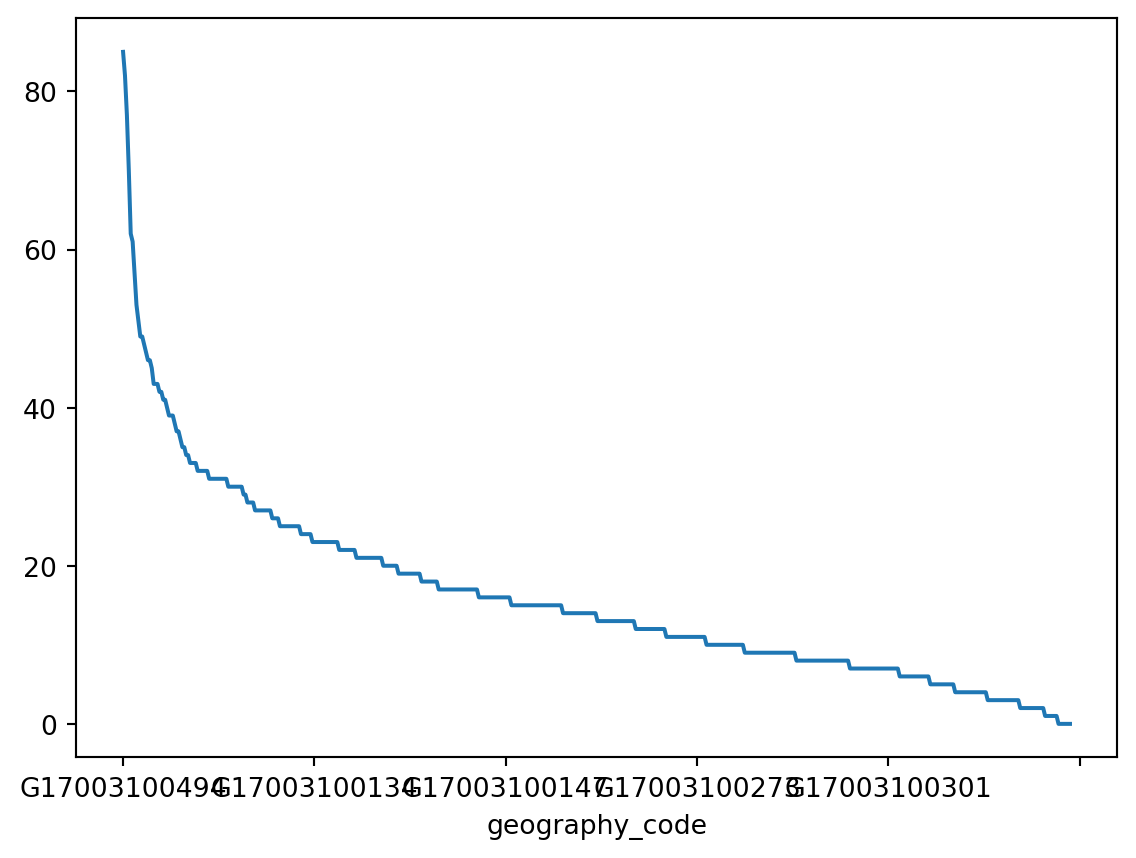
For a bar plot all you need to do is to change from plot to plot.bar. Since there are many census tracts, let us plot only the ten largest ones (which you can retrieve with head):
_ = chicago_1918["influenza"].sort_values(ascending=False).head(10).plot.bar()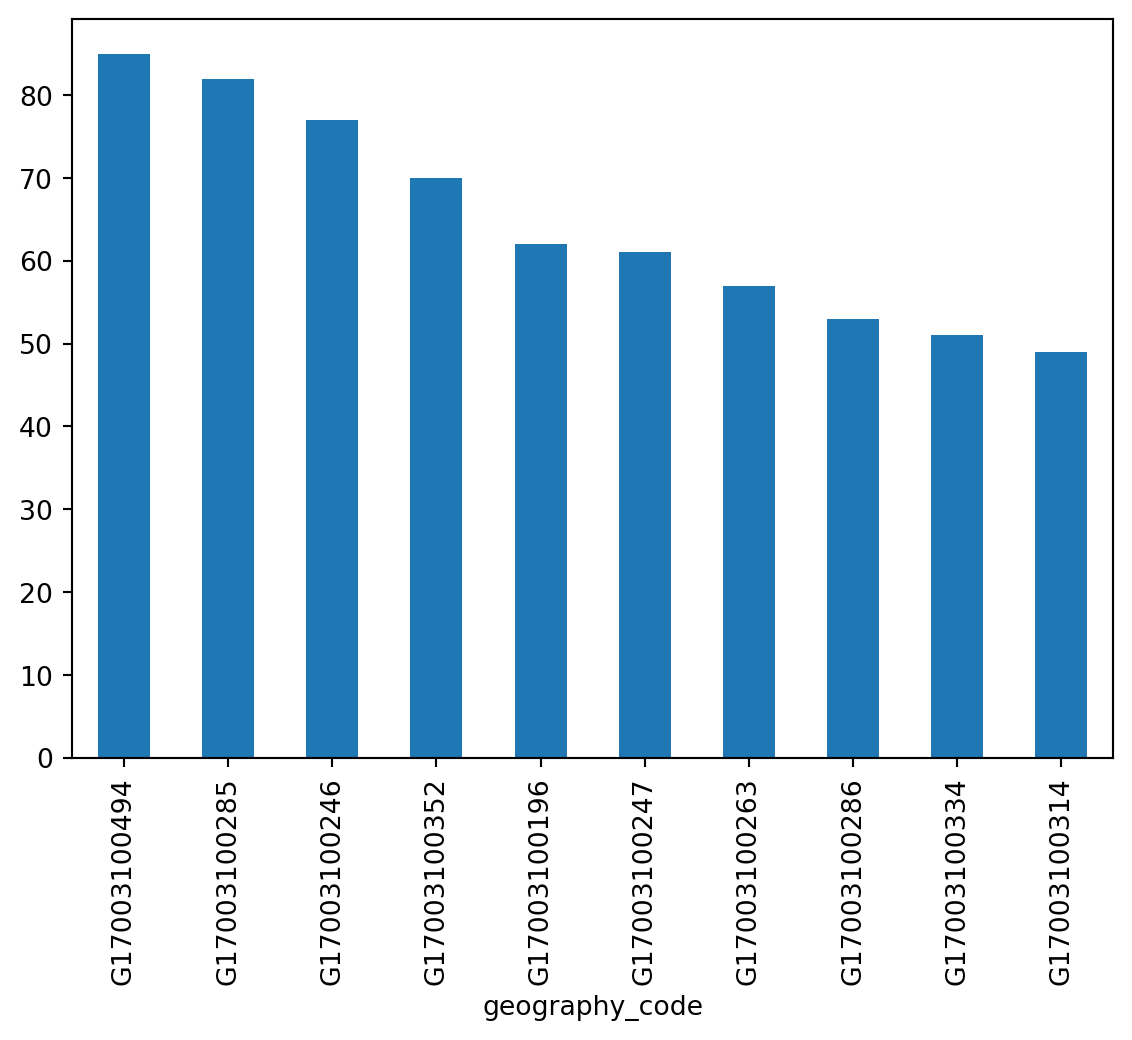
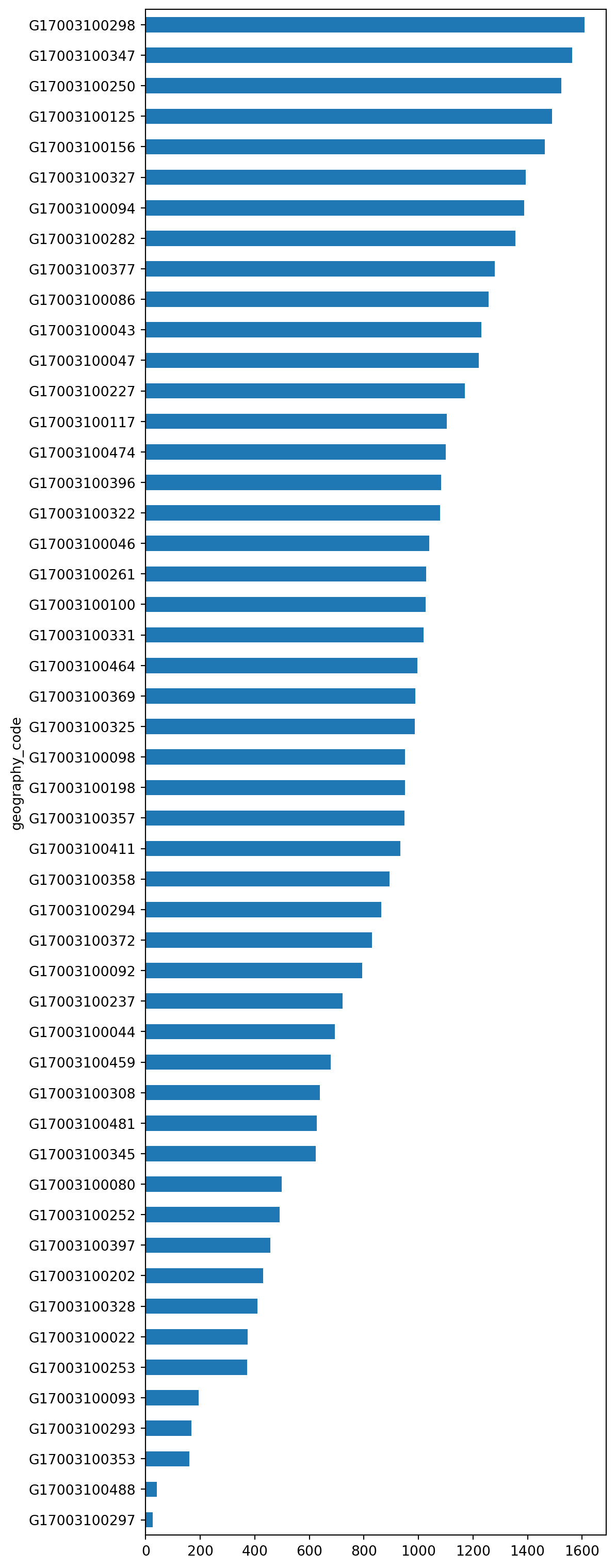
You can turn the plot around by displaying the bars horizontally (see how it’s just changing bar for barh). Let’s display now the top 50 areas and, to make it more readable, let us expand the plot’s height:
_ = (
chicago_1918["total_population"]
.sort_values()
.head(50)
.plot.barh(figsize=(6, 20))
)
You may have noticed that in some cases, the code is on a single line, but longer code is split into multiple lines. Python requires you to follow the indentation rules, but apart from that, there are not a lot of other limits.
This section is a bit more advanced and hence considered optional. Feel free to skip it, move to the next, and return later when you feel more confident.
Once you can read your data in, explore specific cases, and have a first visual approach to the entire set, the next step can be preparing it for more sophisticated analysis. Maybe you are thinking of modeling it through regression, or on creating subgroups in the dataset with particular characteristics, or maybe you simply need to present summary measures that relate to a slightly different arrangement of the data than you have been presented with.
For all these cases, you first need what statistician, and general R wizard, Hadley Wickham calls “tidy data”. The general idea to “tidy” your data is to convert them from whatever structure they were handed in to you into one that allows convenient and standardized manipulation, and that supports directly inputting the data into what he calls “tidy” analysis tools. But, at a more practical level, what is exactly “tidy data”? In Wickham’s own words:
Tidy data is a standard way of mapping the meaning of a dataset to its structure. A dataset is messy or tidy depending on how rows, columns and tables are matched up with observations, variables and types.
He then goes on to list the three fundamental characteristics of “tidy data”:
If you are further interested in the concept of “tidy data”, I recommend you check out the original paper (open access) and the public repository associated with it.
Let us bring in the concept of “tidy data” to our own Chicago dataset. First, remember its structure:
chicago_1918.head()| gross_acres | illit | unemployed_pct | ho_pct | agecat1 | agecat2 | agecat3 | agecat4 | agecat5 | agecat6 | agecat7 | influenza | total_population | flu_rate | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| geography_code | ||||||||||||||
| G17003100001 | 1388.2 | 116 | 0.376950 | 0.124823 | 46 | 274 | 257 | 311 | 222 | 1122 | 587 | 9 | 2819 | 0.003193 |
| G17003100002 | 217.7 | 14 | 0.399571 | 0.071647 | 35 | 320 | 441 | 624 | 276 | 1061 | 508 | 6 | 3265 | 0.001838 |
| G17003100003 | 401.3 | 69 | 0.349558 | 0.092920 | 50 | 265 | 179 | 187 | 163 | 1020 | 392 | 8 | 2256 | 0.003546 |
| G17003100004 | 86.9 | 11 | 0.422535 | 0.030072 | 43 | 241 | 129 | 141 | 123 | 1407 | 539 | 2 | 2623 | 0.000762 |
| G17003100005 | 337.1 | 20 | 0.431822 | 0.084703 | 65 | 464 | 369 | 464 | 328 | 2625 | 1213 | 7 | 5528 | 0.001266 |
Thinking through tidy lenses, this is not a tidy dataset. It is not so for each of the three conditions:
geography_code; and different observatoins for each area. To tidy up this aspect, you can create separate tables. You will probably want population groups divided by age as one tidy table and flu rates as another. Start by extracting relevant columns.1influenza_rates = chicago_1918[["influenza"]]
influenza_rates.head()chicago_1918["influenza"] but a subset of columns. Just that the subset contains only one column, so you pass a list with a single column name as chicago_1918[["influenza"]]. Notice the double brackets.
| influenza | |
|---|---|
| geography_code | |
| G17003100001 | 9 |
| G17003100002 | 6 |
| G17003100003 | 8 |
| G17003100004 | 2 |
| G17003100005 | 7 |
population = chicago_1918.loc[:, "agecat1":"agecat7"]
population.head()| agecat1 | agecat2 | agecat3 | agecat4 | agecat5 | agecat6 | agecat7 | |
|---|---|---|---|---|---|---|---|
| geography_code | |||||||
| G17003100001 | 46 | 274 | 257 | 311 | 222 | 1122 | 587 |
| G17003100002 | 35 | 320 | 441 | 624 | 276 | 1061 | 508 |
| G17003100003 | 50 | 265 | 179 | 187 | 163 | 1020 | 392 |
| G17003100004 | 43 | 241 | 129 | 141 | 123 | 1407 | 539 |
| G17003100005 | 65 | 464 | 369 | 464 | 328 | 2625 | 1213 |
At this point, the table influenza_rates is tidy: every row is an observation, every table is a variable, and there is only one observational unit in the table.
The other table (population), however, is not entirely tidied up yet: there is only one observational unit in the table, true; but every row is not an observation, and there are variable values as the names of columns (in other words, every column is not a variable). To obtain a fully tidy version of the table, you need to re-arrange it in a way that every row is an age category in an area, and there are three variables: geography_code, age category, and population count (or frequency).
Because this is actually a fairly common pattern, there is a direct way to solve it in pandas:
tidy_population = population.stack()
tidy_population.head()geography_code
G17003100001 agecat1 46
agecat2 274
agecat3 257
agecat4 311
agecat5 222
dtype: int64The method stack, well, “stacks” the different columns into rows. This fixes our “tidiness” problems but the type of object that is returning is not a DataFrame:
type(tidy_population)pandas.core.series.SeriesIt is a Series, which really is like a DataFrame, but with only one column. The additional information (geography_code and age category) are stored in what is called an multi-index. You will skip these for now, so you would really just want to get a DataFrame as you know it out of the Series. This is also one line of code away:
tidy_population_df = tidy_population.reset_index()
tidy_population_df.head()| geography_code | level_1 | 0 | |
|---|---|---|---|
| 0 | G17003100001 | agecat1 | 46 |
| 1 | G17003100001 | agecat2 | 274 |
| 2 | G17003100001 | agecat3 | 257 |
| 3 | G17003100001 | agecat4 | 311 |
| 4 | G17003100001 | agecat5 | 222 |
To which you can apply to renaming to make it look better:
tidy_population_df = tidy_population_df.rename(
columns={"level_1": "age_category", 0: "count"}
)
tidy_population_df.head()| geography_code | age_category | count | |
|---|---|---|---|
| 0 | G17003100001 | agecat1 | 46 |
| 1 | G17003100001 | agecat2 | 274 |
| 2 | G17003100001 | agecat3 | 257 |
| 3 | G17003100001 | agecat4 | 311 |
| 4 | G17003100001 | agecat5 | 222 |
Now our table is fully tidied up!
One of the advantage of tidy datasets is they allow to perform advanced transformations in a more direct way. One of the most common ones is what is called “group-by” operations. Originated in the world of databases, these operations allow you to group observations in a table by one of its labels, index, or category, and apply operations on the data group by group.
For example, given our tidy table with age categories, you might want to compute the total sum of the population by each category. This task can be split into two different ones:
count for each of them.To do this in pandas, meet one of its workhorses, and also one of the reasons why the library has become so popular: the groupby operator.
pop_grouped = tidy_population_df.groupby("age_category")
pop_grouped<pandas.core.groupby.generic.DataFrameGroupBy object at 0x7f070be1e570>The object pop_grouped still hasn’t computed anything. It is only a convenient way of specifying the grouping. But this allows us then to perform a multitude of operations on it. For our example, the sum is calculated as follows:
1pop_grouped.sum(numeric_only=True)numeric_only=False to see the difference.
| count | |
|---|---|
| age_category | |
| agecat1 | 50776 |
| agecat2 | 275363 |
| agecat3 | 201654 |
| agecat4 | 259954 |
| agecat5 | 206358 |
| agecat6 | 1171345 |
| agecat7 | 522130 |
Similarly, you can also obtain a summary of each group:
pop_grouped.describe()| count | ||||||||
|---|---|---|---|---|---|---|---|---|
| count | mean | std | min | 25% | 50% | 75% | max | |
| age_category | ||||||||
| agecat1 | 496.0 | 102.370968 | 78.677423 | 0.0 | 46.75 | 82.0 | 136.00 | 427.0 |
| agecat2 | 496.0 | 555.167339 | 423.526444 | 3.0 | 256.50 | 442.5 | 717.50 | 2512.0 |
| agecat3 | 496.0 | 406.560484 | 301.564896 | 1.0 | 193.50 | 331.5 | 532.50 | 1917.0 |
| agecat4 | 496.0 | 524.100806 | 369.875444 | 4.0 | 253.75 | 453.5 | 709.50 | 2665.0 |
| agecat5 | 496.0 | 416.044355 | 281.825682 | 0.0 | 220.50 | 377.0 | 551.75 | 2454.0 |
| agecat6 | 496.0 | 2361.582661 | 1545.469426 | 8.0 | 1169.75 | 2102.0 | 3191.75 | 9792.0 |
| agecat7 | 496.0 | 1052.681452 | 722.955717 | 6.0 | 519.75 | 918.5 | 1379.25 | 4163.0 |
You will not get into it today as it goes beyond the basics this session wants to cover, but keep in mind that groupby allows you to not only call generic functions (like sum or describe), but also your own functions. This opens the door for virtually any kind of transformation and aggregation possible.
seaborn is an excellent choice. Its online tutorial is a fantastic place to start.This section is derived from A Course on Geographic Data Science by Arribas-Bel (2019), licensed under CC-BY-SA 4.0. The text was slightly adapted, mostly to accommodate a different dataset used.